name: title layout: true class: center --- layout: false count: false .center[ <a href="https://www.nipreps.org/assets/fmriprep-bootcamp-geneva2024/day1-02-bids/"> <img src="images/qr-talk-url.svg" alt="workflow" style="width: 20%" /> <br /> https://www.nipreps.org/assets/fmriprep-bootcamp-geneva2024/day1-02-bids/ </a> <br /> <br /> ## The Brain Imaging Data Structure (BIDS) Chris Markiewicz <<code>markiewicz@stanford.edu</code>> Oscar Esteban <<code>phd@oscaresteban.es</code>> ] ??? NOTES TO OURSELVES GO HERE --- name: newsection layout: true .perma-sidebar[ <p class="rotate"> <a rel="license" href="http://creativecommons.org/licenses/by/4.0/"><img alt="Creative Commons License" style="border-width:0; height: 20px; padding-top: 6px;" src="https://i.creativecommons.org/l/by/4.0/88x31.png" /></a> <span style="padding-left: 10px; font-weight: 600;">Day 1 :: Brain Imaging Data Structure</span> </p> ] --- <br /> <br /> <br /> <br /> <br /> <br /> .boxed-content[ .large.center[ .larger.center.gray-text[Before we start...] ## What do you think/know about BIDS? ]] --- layout: true template: newsection name: BIDS # BIDS: Brain Imaging Data Structure .left-column2.small[ .pad-left[ <i class="fa-solid fa-folder-open"></i> my_dataset/<br /> .pad-left[<i class="fa-solid fa-file-lines"></i> CHANGES]<br /> .pad-left[<i class="fa-solid fa-file-lines"></i> README]<br /> .pad-left[<i class="fa-solid fa-paperclip"></i> dataset_description.json]<br /> .pad-left[<i class="fa-solid fa-paperclip"></i> participants.json]<br /> .pad-left[<i class="fa-solid fa-table"></i> participants.tsv]<br /> .pad-left[<i class="fa-solid fa-paperclip"></i> scans.json]<br /> .pad-left[<i class="fa-solid fa-folder-open"></i> sub-11]<br /> .pad-left[.pad-left[<i class="fa-solid fa-folder-open"></i> anat/]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-paperclip"></i> sub-11_T2w.json]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_T2w.nii.gz]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-paperclip"></i> sub-11_acq-denoised_T1w.json]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_acq-denoised_T1w.nii.gz]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-paperclip"></i> sub-11_acq-mp2rage_T1w.json]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_acq-mp2rage_T1w.nii.gz]]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-folder-open"></i> fmap/]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-paperclip"></i> sub-11_magnitude1.json]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_magnitude1.nii.gz]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-paperclip"></i> sub-11_magnitude2.json]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_magnitude2.nii.gz]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-paperclip"></i> sub-11_phasediff.json]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_phasediff.nii.gz]]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-folder-open"></i> func/]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-paperclip"></i> sub-11_task-mixed_bold.json]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_task-mixed_bold.nii.gz]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-table"></i> sub-11_task-mixed_events.tsv]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-paperclip"></i> sub-11_task-rest_bold.json]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_task-rest_bold.nii.gz]]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-table"></i> sub-11_scans.tsv]]<br /> .pad-left[<i class="fa-solid fa-folder"></i> sub-14]<br /> .pad-left[<i class="fa-solid fa-folder"></i> sub-15]<br /> ] ] --- .right-column2.larger[ * BIDS is a directory structure, based on common practices * Builds on existing standards (NIfTI, JSON, TSV) * Intended for human *and* machine legibility * The [BIDS Validator](https://bids-standard.github.io/bids-validator) makes compliance easy to verify * The [specification](https://bids-specification.readthedocs.io/en/stable/) is a searchable HTML document ] --- .right-column2[.no-bullet[* .larger[Basic metadata in the file names] * Subject, session, imaging modality, etc. * Generally just enough to assign unique names]] -- .right-column2[.no-bullet[* .larger[NIfTI headers and *JSON sidecars* contain detailed, image-related metadata]]] -- .right-column2[.no-bullet[* .larger[Study-level metadata] * Not necessarily associated with specific files * [`dataset_description.json`](https://bids-specification.readthedocs.io/en/stable/03-modality-agnostic-files.html#dataset_descriptionjson), [`participants.tsv`](https://bids-specification.readthedocs.io/en/stable/03-modality-agnostic-files.html#participants-file), [`sessions.tsv`](https://bids-specification.readthedocs.io/en/stable/05-longitudinal-and-multi-site-studies.html#sessions-file), and [`scans.tsv`](https://bids-specification.readthedocs.io/en/stable/03-modality-agnostic-files.html#scans-file) ]] --- template: newsection layout: true --- <br /> .boxed-content[ <figure style="width: 100%">  </figure> ] --- # A staggering pace of adoption <br /> .boxed-content[ <figure style="width: 100%">  </figure> ] --- # bids-specification.readthedocs.io .boxed-content[ <iframe src="https://bids-specification.readthedocs.io/en/stable/" width="100%" height="540px" style="border: 0; margin-top: 15px" /> ] --- # More reproducible research .boxed-content[ .no-bullet[ * .larger[<i class="fa-solid fa-arrow-right-arrow-left"></i> BIDS defines a clear interface] * Standardized through a large-community agreement * Human readable (as opposed to DICOM) * Machine readable (enabling tooling, see below) * Stable (e.g., backward compatibility principle), extensible, and predictable (*BIDS Schema*) * .larger[<i class="fa-solid fa-box-archive"></i> Harnesses data-sharing and archival (sharing with self)] * Can be checked for correctness (*BIDS Validator*) * Establishes a solid *contract* on the organization of data between authors and reusers (or your future self) * Separates *data blobs* and metadata, facilitating agile management and understanding of the dataset, HPC operations (e.g., *datalad drop*), etc. * Enables open-sharing (e.g., *OpenNeuro*) * .larger[<i class="fa-solid fa-gear"></i> Supports tooling] * Querying the dataset, filtering data, etc. (*PyBIDS*) * Conversion from other formats (mostly *DICOM*) * Interpreting unprocessed data (development of *BIDS Apps* such as *MRIQC* or *fMRIPrep*) * *BIDS Derivatives* enable concatenating *BIDS Apps* (e.g., *fMRIPrep* + *XCPD*, *brainlife.io*) * Data-management-friendly (*DataLad*) ] ] --- # BIDS Validator .boxed-content[ The machine-readable structure makes validation possible: ``` bash $ bids-validator my_dataset/ bids-validator@1.14.6 1: [WARN] The recommended file /README is very small. Please consider expanding it with additional information about the dataset. (code: 213 - README_FILE_SMALL) ./README Please visit https://neurostars.org/search?q=README_FILE_SMALL for existing conversations about this issue. Summary: Available Tasks: Available Modalities: 96 Files, 6.63GB Mixed-stimuli, positive-control task MRI 5 - Subjects Resting-state fMRI 1 - Session If you have any questions, please post on https://neurostars.org/tags/bids. ``` .center[ ## https://bids-standard.github.io/bids-validator/ ]] --- # BIDS Validator .boxed-content[ Once all warnings and errors are addressed: ``` bash $ bids-validator my_dataset/ bids-validator@1.14.6 This dataset appears to be BIDS compatible. Summary: Available Tasks: Available Modalities: 96 Files, 6.63GB Mixed-stimuli, positive-control task MRI 5 - Subjects Resting-state fMRI 1 - Session If you have any questions, please post on https://neurostars.org/tags/bids. ``` .center[ ## https://bids-standard.github.io/bids-validator/ ]] --- # Sharing through OpenNeuro .boxed-content[ .center[[https://openneuro.org/datasets/ds005454/versions/1.0.0](https://openneuro.org/datasets/ds005454/versions/1.0.0)] <iframe src="https://openneuro.org/datasets/ds005454/versions/1.0.0" width="100%" height="500px" style="border: 0" /> ] --- layout: true template: newsection name: Apps # BIDS Applications --- A common specification of neuroimaging datasets affords queries for and adaptation to the available data. -- .boxed-content[ Queryable (meta)data allows a very simple protocol for a [BIDS App](https://bids-apps.neuroimaging.io/apps/): ```Bash bids-app /bids-directory /output-directory participant [OPTIONS] ``` .footnote[ \* Note that `participant` is an analysis level. Apps may also operate at the `run`, `session` or `group` levels. ]] -- .boxed-content[ ## Examples **MRIQC** ```Bash mriqc /data/bids/openneuro/ds000228 /data/processed/ds000228-mriqc group ``` ] -- .boxed-content[ **fMRIPrep** ```Bash fmriprep /data/bids/openneuro/ds000228 /data/processed/ds000228-fmriprep \ participant --participant-label pixar001 ``` ] --- .pull-left[ ### Many application types are possible ] .pull-right[ [](https://bids-apps.neuroimaging.io/apps/) ] --- count: false .pull-left[ ### Many application types are possible * Quality control ] .pull-right[ [](https://bids-apps.neuroimaging.io/apps/) ] --- count: false .pull-left[ ### Many application types are possible * Quality control * Anatomical pipelines ] .pull-right[ [](https://bids-apps.neuroimaging.io/apps/) ] --- count: false .pull-left[ ### Many application types are possible * Quality control * Anatomical pipelines * Functional pipelines ] .pull-right[ [](https://bids-apps.neuroimaging.io/apps/) ] --- count: false .pull-left[ ### Many application types are possible * Quality control * Anatomical pipelines * Functional pipelines * Diffusion pipelines ] .pull-right[ [](https://bids-apps.neuroimaging.io/apps/) ] --- count: false .pull-left[ ### Many application types are possible * Quality control * Anatomical pipelines * Functional pipelines * Diffusion pipelines ### Lowered friction encourages adoption * Researchers gain easy access to tools by formatting data in BIDS * Accepting BIDS datasets makes your tools easy to try ] .pull-right[ [](https://bids-apps.neuroimaging.io/apps/) ] --- template: newsection layout: true name: Derivatives # BIDS Derivatives --- BIDS Derivatives generally mirror the raw dataset. .small[ .pad-left[ <i class="fa-solid fa-folder-open"></i> fmriprep/<br /> .pad-left[<i class="fa-solid fa-file-lines"></i> README]<br /> .pad-left[<i class="fa-solid fa-circle-info"></i> dataset_description.json]<br /> .pad-left[<i class="fa-solid fa-folder-open"></i> sub-11]<br /> .pad-left[.pad-left[<i class="fa-solid fa-folder-open"></i> anat/]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_desc-preproc_T2w.nii.gz]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_acq-denoised_desc-preproc_T1w.nii.gz]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_acq-denoised_desc-brain_mask.nii.gz]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_acq-denoised_from-T1w_to-MNI152NLin2009cAsym_mode-image_xfm.h5]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-blank fa-brain"></i> ...]]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-folder-open"></i> fmap/]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_fmapid-auto00000_desc-coeff_fieldmap.nii.gz]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_fmapid-auto00000_desc-magnitude_fieldmap.nii.gz]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_fmapid-auto00000_desc-preproc_fieldmap.nii.gz]]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-folder-open"></i> func/]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_task-mixed_desc-coreg_boldref.nii.gz]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_task-mixed_desc-hmc_boldref.nii.gz]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-file-lines"></i> sub-11_task-mixed_from-boldref_to-T1w_mode-image_desc-coreg_xfm.txt]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-file-lines"></i> sub-11_task-mixed_from-boldref_to-auto00000_mode-image_desc-fmapreg_xfm.txt]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_task-mixed_from-orig_to-boldref_mode-image_desc-hmc_xfm.txt]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_task-mixed_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-blank fa-brain"></i> ...]]]<br /> .pad-left[<i class="fa-solid fa-folder"></i> sub-14]<br /> .pad-left[<i class="fa-solid fa-folder"></i> sub-15]<br /> ] ] -- New file suffixes, extensions and entities are possible. --- template: newsection layout: true name: PyBIDS .install-cmd[ ```Bash pip install pybids ``` ] --- # PyBIDS .boxed-content[ A common specification of neuroimaging datasets affords queries for and adaptation to the available data. [PyBIDS](https://github.com/bids-standard/pybids/) is a Python library for querying and manipulating BIDS datasets. ```Python >>> from bids import BIDSLayout >>> layout = BIDSLayout('/my_dataset') >>> bold = layout.get(subject='11', task='rest', suffix='bold', extension='.nii.gz') >>> bold [<BIDSImageFile filename='/my_dataset/sub-11/func/sub-11_task-rest_bold.nii.gz'>] >>> md = bold[0].get_metadata() >>> md['RepetitionTime'] 1.5 >>> bold[0].get_entities() {'datatype': 'func', 'extension': '.nii.gz', 'subject': '11', 'suffix': 'bold', 'task': 'rest'} ``` ] --- # PyBIDS | Derivatives .boxed-content[ PyBIDS layouts support BIDS derivatives: ```Python >>> fmri_derivs = BIDSLayout( ... '/my_dataset-fmriprep', ... config=['bids', 'derivatives'], ... validate=False, ... ) >>> spaces = fmri_derivs.get_spaces(desc='preproc', suffix='bold') >>> spaces ['MNI152NLin2009cAsym'] ``` ] -- .boxed-content[ It is also possible to provide your own config: ```Python >>> import niworkflows >>> nipreps_config = niworkflows.data.load('nipreps.json') >>> fmri_derivs = BIDSLayout('/my_dataset-fmriprep', config=[nipreps_config], validate=False) ``` ] --- # PyBIDS | Large datasets .boxed-content[ <br /> .larger[*BIDS Apps* can spend long time indexing if they rely on *PyBIDS* ] .no-bullet[ * <i class="fa-solid fa-circle-right"></i> **create a cache**: ] ```Bash $ pybids layout --reset-db --no-validate --index-metadata . /my_dataset/.bids-index/ ``` .larger[And then inform your *BIDS App*.] <br /> In the case of *MRIQC* and *fMRIPrep*, point the `--bids-database-dir` argument to the newly created cache: ```Bash $ mriqc /data/input /derivatives/output participant [OPTIONS] \ --bids-database-dir /my_dataset/.bids-index/ ``` ```Bash $ fmriprep /data/input /derivatives/output participant [OPTIONS] \ --bids-database-dir /my_dataset/.bids-index/ ``` ] --- template: newsection layout: true name: conversion --- # Converting from DICOM into BIDS .left-column-mid.small[ .pad-left[ <i class="fa-solid fa-folder-open"></i> my_dataset/<br /> .pad-left[<i class="fa-solid fa-folder-open"></i> sub-11]<br /> .pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 10 - anat-T1w_acq-mp2rage_run-01_UNI_Images.dicom]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 11 - anat-T1w_acq-mp2rage_run-01_UNI-DEN.dicom]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 12 - anat-T1w_acq-mp2rage_run-01_INV2.dicom]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-folder-open"></i> 16 - fmap-gre_acq-siemens_run-01/]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 1.3.12.2.1107.5.2.61.237203.20[...]25.MR.dcm]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 1.3.12.2.1107.5.2.61.237203.20[..]33.MR.dcm]]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 17 - fmap-gre_acq-siemens_run-01.dicom ]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-folder-open"></i> 18 - func-bold_acq-midRes_task-rest_run-01/ ]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 1.3.12.2.1107.5.2.61.237203.20[...]25.MR.dcm]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-ellipsis"></i>]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 1.3.12.2.1107.5.2.61.237203.20[..]33.MR.dcm]]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-folder-open"></i> 20 - func-bold_acq-midRes_task-mixed_run-01/ ]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 1.3.12.2.1107.5.2.61.237203.20[...]25.MR.dcm]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-ellipsis"></i>]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 1.3.12.2.1107.5.2.61.237203.20[..]33.MR.dcm]]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 22 - anat-T2w_acq-p6_run-01.dicom]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 7 - anat-T1w_acq-mp2rage_run-01_INV1.dicom]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 8 - anat-T1w_acq-mp2rage_run-01_T1_Images.dicom]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 9 - anat-T1w_acq-mp2rage_run-01_T1_Images.dicom]]<br /> .pad-left[<i class="fa-solid fa-folder"></i> sub-14]<br /> .pad-left[<i class="fa-solid fa-folder"></i> sub-15]<br /> ] ] .right-column-mid[ .no-bullet[ * .larger[DICOM (Digital Imaging and Communications in Medicine)] * was originally meant for storage and transmission within/between machines. * .larger[DICOM structures store their relevant metadata internally,] * but structure and fields tend to be vendor- & institution-specific. * .larger[Constructing a BIDS dataset can be tedious and error-prone, so there are several [converters](https://bids.neuroimaging.io/benefits.html#converters).] * The [Heuristic DICOM Converter](https://heudiconv.readthedocs.io/) (HeuDiConv) and [BIDSCoin](https://bidscoin.readthedocs.io/en/stable/) allow you to specify the mapping and apply in bulk. Both build off of [dcm2niix](https://github.com/rordenlab/dcm2niix/). ] ] --- # Converting from DICOM into BIDS .left-column-mid.small[ .pad-left[ <i class="fa-solid fa-folder-open"></i> my_dataset/<br /> .pad-left[<i class="fa-solid fa-folder-open"></i> sub-11]<br /> .pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 10 - anat-T1w_acq-mp2rage_run-01_UNI_Images.dicom]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 11 - anat-T1w_acq-mp2rage_run-01_UNI-DEN.dicom]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 12 - anat-T1w_acq-mp2rage_run-01_INV2.dicom]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-folder-open"></i> 16 - fmap-gre_acq-siemens_run-01/]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 1.3.12.2.1107.5.2.61.237203.20[...]25.MR.dcm]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 1.3.12.2.1107.5.2.61.237203.20[..]33.MR.dcm]]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 17 - fmap-gre_acq-siemens_run-01.dicom ]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-folder-open"></i> 18 - func-bold_acq-midRes_task-rest_run-01/ ]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 1.3.12.2.1107.5.2.61.237203.20[...]25.MR.dcm]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-ellipsis"></i>]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 1.3.12.2.1107.5.2.61.237203.20[..]33.MR.dcm]]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-folder-open"></i> 20 - func-bold_acq-midRes_task-mixed_run-01/ ]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 1.3.12.2.1107.5.2.61.237203.20[...]25.MR.dcm]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-ellipsis"></i>]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 1.3.12.2.1107.5.2.61.237203.20[..]33.MR.dcm]]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 22 - anat-T2w_acq-p6_run-01.dicom]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 7 - anat-T1w_acq-mp2rage_run-01_INV1.dicom]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 8 - anat-T1w_acq-mp2rage_run-01_T1_Images.dicom]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 9 - anat-T1w_acq-mp2rage_run-01_T1_Images.dicom]]<br /> .pad-left[<i class="fa-solid fa-folder"></i> sub-14]<br /> .pad-left[<i class="fa-solid fa-folder"></i> sub-15]<br /> ] ] .right-column-mid.small[ .pad-left[ <i class="fa-solid fa-folder-open"></i> my_dataset/<br /> .pad-left[<i class="fa-solid fa-file-lines"></i> CHANGES]<br /> .pad-left[<i class="fa-solid fa-file-lines"></i> README]<br /> .pad-left[<i class="fa-solid fa-paperclip"></i> dataset_description.json]<br /> .pad-left[<i class="fa-solid fa-paperclip"></i> participants.json]<br /> .pad-left[<i class="fa-solid fa-table"></i> participants.tsv]<br /> .pad-left[<i class="fa-solid fa-paperclip"></i> scans.json]<br /> .pad-left[<i class="fa-solid fa-folder-open"></i> sub-11]<br /> .pad-left[.pad-left[<i class="fa-solid fa-folder-open"></i> anat/]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-paperclip"></i> sub-11_T2w.json]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_T2w.nii.gz]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-paperclip"></i> sub-11_acq-denoised_T1w.json]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_acq-denoised_T1w.nii.gz]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-paperclip"></i> sub-11_acq-mp2rage_T1w.json]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_acq-mp2rage_T1w.nii.gz]]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-folder-open"></i> fmap/]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-paperclip"></i> sub-11_magnitude1.json]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_magnitude1.nii.gz]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-paperclip"></i> sub-11_magnitude2.json]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_magnitude2.nii.gz]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-paperclip"></i> sub-11_phasediff.json]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_phasediff.nii.gz]]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-folder-open"></i> func/]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-paperclip"></i> sub-11_task-mixed_bold.json]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_task-mixed_bold.nii.gz]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-table"></i> sub-11_task-mixed_events.tsv]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-paperclip"></i> sub-11_task-rest_bold.json]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_task-rest_bold.nii.gz]]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-table"></i> sub-11_scans.tsv]]<br /> .pad-left[<i class="fa-solid fa-folder"></i> sub-14]<br /> .pad-left[<i class="fa-solid fa-folder"></i> sub-15]<br /> ] ] <i class="fa-solid fa-circle-right" style="margin-top: 20%; font-size: 1.7em"></i> --- # DICOM-to-BIDS: ReproIn .center[[ReproIn](https://github.com/ReproNim/reproin) standardizes protocol naming conventions:] <figure style="width: 60%"> 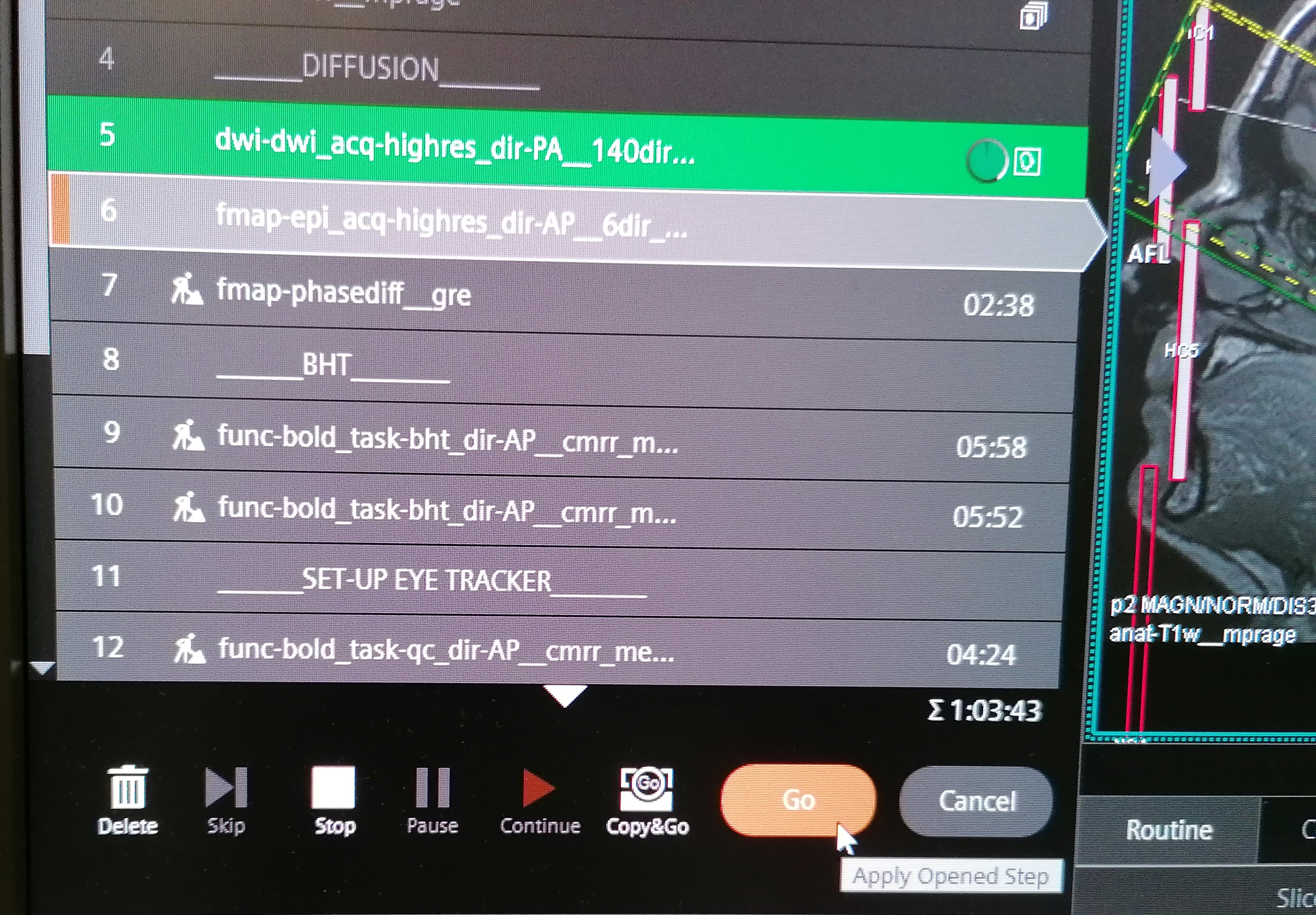 <figcaption>From <a href="https://www.axonlab.org/hcph-sops">the HCPh SOPs</a>; doi:</figcaption> </figure> --- <figure style="width: 60%">  <figcaption>From <a href="https://github.com/ReproNim/reproin/blob/master/README.md#overall-workflow">ReproIn</a>; doi:</figcaption> ??? Please note that this figure is OUT OF DATE. For example, the anatomicals are called anat-T1W__, anat-T2w__ etc. --- # DICOM-to-BIDS: HeuDiConv .boxed-content.no-bullet[ * .large[<i class="fa-solid fa-circle-chevron-right"></i> **Heu**ristic **DI**COM** Conv**erter: maps DICOM metadata into BIDS] * .large[<i class="fa-solid fa-circle-chevron-right"></i> Mappings are encoded within the *heuristic file*] * The `.heudiconv/` folder contains the metadata that can be employed in the *heuristic file* to define mappings.<br /><br /> <figure style="width: 100%"> 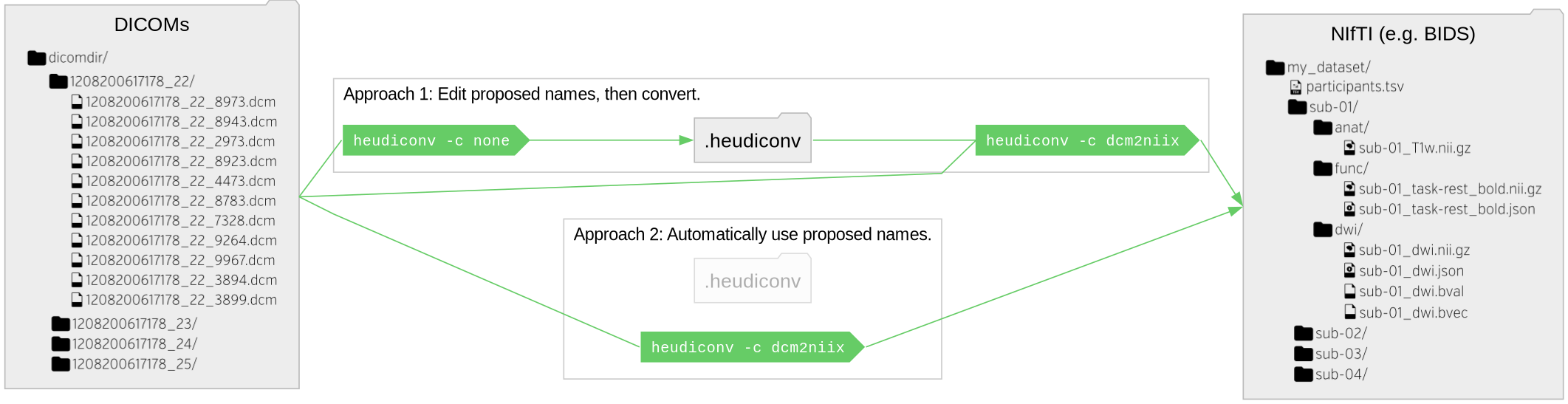 <figcaption>From <a href="https://github.com/nipy/heudiconv?tab=readme-ov-file#howto-101">HeuDiConv</a>; doi:</figcaption> ] --- # DICOM-to-BIDS: HeuDiConv .boxed-content[ .no-bullet.large[ * <i class="fa-solid fa-1"></i> Generate a **heuristic file** * <i class="fa-solid fa-2"></i> Edit the **heuristic file** ] .small[ ```Python data = create_key('run-{item:03d}') t1w = create_key('sub-{subject}/{session}/anat/sub-{subject}_{session}_T1w') dwi = create_key('sub-{subject}/{session}/dwi/sub-{subject}_{session}_dir-AP_dwi') # Save the RPE (reverse phase-encode) B0 image as a fieldmap (fmap). It will be used to correct # the distortion in the DWI fmap_rev_phase = create_key('sub-{subject}/{session}/fmap/sub-{subject}_{session}_dir-PA_epi') fmap_mag = create_key('sub-{subject}/{session}/fmap/sub-{subject}_{session}_magnitude') fmap_phase = create_key('sub-{subject}/{session}/fmap/sub-{subject}_{session}_phasediff') # Even if this is resting state, you still need a task key func_rest = create_key('sub-{subject}/{session}/func/sub-{subject}_{session}_task-rest_run-01_bold') func_rest_post = create_key('sub-{subject}/{session}/func/sub-{subject}_{session}_task-rest_run-02_bold') ``` ] .no-bullet.large[ * <i class="fa-solid fa-3"></i> Execute *HeuDiConv* ] ] --- # DICOM-to-BIDS: ReproIn + HeuDiConv .no-bullet.large[ * <i class="fa-solid fa-circle-chevron-right"></i> Just run *HeuDiConv* directly! *Heuristic file* is pre-defined. * <i class="fa-solid fa-circle-question"></i> What if I **did not** follow *ReproIn* namings? ] -- .boxed-content.small[ ``` Python # A dictionary containing fixes/remapping for sequence names per study. # Keys are md5sum of study_description from DICOMs, in the form of PI-Experimenter^protocolname # You can use `heudiconv -f reproin --command ls --files PATH # to list the "study hash". # Values are list of tuples in the form (regex_pattern, substitution). # If the key is an empty string "", it would apply to any study. protocols2fix: dict[str | re.Pattern[str], list[tuple[str, str]]] = { "": [ # <-- empty string means apply to whole study ("anat-T1w_acq-mp2rage_run-01", "anat-mp2rage"), ("_UNI_", "_acq-mp2rage__"), ("_UNI-DEN", "_acq-denoised__"), ("_INV", "_inv-"), ("_T1_", "_acq-T1map__"), ("fmap_acq-siemens", "fmap-phasediff"), ("fmap-gre_acq-siemens", "fmap-phasediff"), ("_acq-midRes", ""), ("_acq-p6", ""), ("_run-01", ""), ] } ``` ] --- # DICOM-to-BIDS: ReproIn + HeuDiConv .left-column-mid[ Our dataset's DICOM had some non-compliant names: .small[ .pad-left[ <i class="fa-solid fa-folder-open"></i> my_dataset/<br /> .pad-left[<i class="fa-solid fa-folder-open"></i> sub-11]<br /> .pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 10 - anat-T1w_acq-mp2rage_run-01<mark>_UNI_Images</mark>.dicom]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 11 - anat-T1w_acq-mp2rage_run-01<mark>_UNI-DEN</mark>.dicom]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 12 - anat-T1w_acq-mp2rage_run-01<mark>_INV2</mark>.dicom]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-folder-open"></i> 16 - fmap-gre_acq-siemens_run-01/]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 1.3.12.2.1107.5.2.61.237203.20[...]25.MR.dcm]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 1.3.12.2.1107.5.2.61.237203.20[..]33.MR.dcm]]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 17 - <mark>fmap-gre</mark>_acq-siemens_run-01.dicom ]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-folder-open"></i> 18 - func-bold_acq-midRes_task-rest_run-01/ ]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 1.3.12.2.1107.5.2.61.237203.20[...]25.MR.dcm]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-ellipsis"></i>]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 1.3.12.2.1107.5.2.61.237203.20[..]33.MR.dcm]]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-folder-open"></i> 20 - func-bold_acq-midRes_task-mixed_run-01/ ]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 1.3.12.2.1107.5.2.61.237203.20[...]25.MR.dcm]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-ellipsis"></i>]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 1.3.12.2.1107.5.2.61.237203.20[..]33.MR.dcm]]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 22 - anat-T2w_acq-p6_run-01.dicom]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 7 - anat-T1w_acq-mp2rage_run-01<mark>_INV1</mark>.dicom]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 8 - anat-T1w_acq-mp2rage_run-01<mark>_T1_Images</mark>.dicom]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-layer-group"></i> 9 - anat-T1w_acq-mp2rage_run-01<mark>_T1_Images</mark>.dicom]]<br /> .pad-left[<i class="fa-solid fa-folder"></i> sub-14]<br /> .pad-left[<i class="fa-solid fa-folder"></i> sub-15]<br /> ] ]] .right-column-mid.small[ .pad-left[ <i class="fa-solid fa-folder-open"></i> my_dataset/<br /> .pad-left[<i class="fa-solid fa-file-lines"></i> CHANGES]<br /> .pad-left[<i class="fa-solid fa-file-lines"></i> README]<br /> .pad-left[<i class="fa-solid fa-paperclip"></i> dataset_description.json]<br /> .pad-left[<i class="fa-solid fa-paperclip"></i> participants.json]<br /> .pad-left[<i class="fa-solid fa-table"></i> participants.tsv]<br /> .pad-left[<i class="fa-solid fa-paperclip"></i> scans.json]<br /> .pad-left[<i class="fa-solid fa-folder-open"></i> sub-11]<br /> .pad-left[.pad-left[<i class="fa-solid fa-folder-open"></i> anat/]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-paperclip"></i> sub-11_T2w.json]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_T2w.nii.gz]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-paperclip"></i> sub-11_acq-denoised_T1w.json]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_acq-denoised_T1w.nii.gz]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-paperclip"></i> sub-11_acq-mp2rage_T1w.json]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_acq-mp2rage_T1w.nii.gz]]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-folder-open"></i> fmap/]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-paperclip"></i> sub-11_magnitude1.json]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_magnitude1.nii.gz]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-paperclip"></i> sub-11_magnitude2.json]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_magnitude2.nii.gz]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-paperclip"></i> sub-11_phasediff.json]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_phasediff.nii.gz]]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-folder-open"></i> func/]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-paperclip"></i> sub-11_task-mixed_bold.json]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_task-mixed_bold.nii.gz]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-table"></i> sub-11_task-mixed_events.tsv]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-paperclip"></i> sub-11_task-rest_bold.json]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_task-rest_bold.nii.gz]]]<br /> .pad-left[.pad-left[<i class="fa-solid fa-table"></i> sub-11_scans.tsv]]<br /> .pad-left[<i class="fa-solid fa-folder"></i> sub-14]<br /> .pad-left[<i class="fa-solid fa-folder"></i> sub-15]<br /> ] ] <i class="fa-solid fa-circle-right" style="margin-top: 20%; font-size: 1.7em"></i> --- # DICOM-to-BIDS: HeuDiConv may not be sufficient .boxed-content[ .no-bullet[ * .large[<i class="fa-solid fa-timeline"></i> **'events'** files describing the experiment's timeline] ] .pad-left[ <i class="fa-solid fa-folder-open"></i> my_dataset/<br /> .pad-left[<i class="fa-solid fa-folder-open"></i> sub-11]<br /> .pad-left[.pad-left[<i class="fa-solid fa-folder-open"></i> func/]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-paperclip"></i> sub-11_task-mixed_bold.json]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_task-mixed_bold.nii.gz]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-table"></i> <mark>sub-11_task-mixed_events.tsv</mark>]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-paperclip"></i> sub-11_task-rest_bold.json]]]<br /> .pad-left[.pad-left[.pad-left[<i class="fa-solid fa-brain"></i> sub-11_task-rest_bold.nii.gz]]]<br /> ] <br /> .no-bullet[ * .large[<i class="fa-solid fa-heart-pulse"></i> **'physio'** files (physiological recordings, eye-tracking, etc.)] * .large[<i class="fa-solid fa-film"></i> **'stim'** files (stimuli-related such as movies, task cues, etc.)] * .large[<i class="fa-solid fa-suitcase-medical"></i> supporting metadata (e.g., field maps, covariates, phenotype, etc.)] ] ] --- # Conclusion .boxed-content[ .distribute.large[ ] ] ??? --- layout: false count: false .center[ <a href="https://www.nipreps.org/assets/fmriprep-bootcamp-geneva2024/day1-02-bids/"><img src="images/qr-talk-url.svg" alt="workflow" style="width: 20%" /></a> ## Thanks <br /> <br /> #### Chris Markiewicz <<code>markiewicz@stanford.edu</code>> & Oscar Esteban <<code>phd@oscaresteban.es</code>> The Brain Imaging Data Structure (BIDS) Funding: [SNSF 185872](https://data.snf.ch/grants/grant/185872), [RF1MH121867](https://reporter.nih.gov/project-details/10260312), [CZI EOSS5-000266](https://chanzuckerberg.com/eoss/proposals/nipreps-a-community-framework-for-reproducible-neuroimaging/) ] ???