smriprep.workflows.surfaces module
Surface preprocessing workflows.
sMRIPrep uses FreeSurfer to reconstruct surfaces from T1w/T2w structural images.
- smriprep.workflows.surfaces.init_anat_ribbon_wf(name='anat_ribbon_wf')[source]
Create anatomical ribbon mask
Workflow Graph
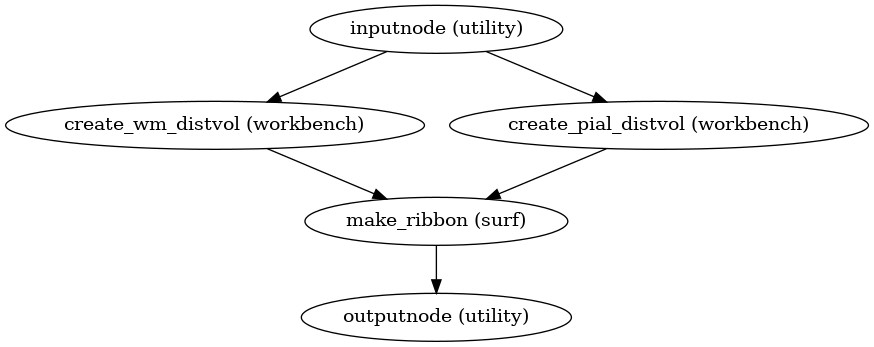
(Source code, png, svg, pdf)
- Inputs:
white – Left and right gray/white surfaces (as GIFTI files)
pial – Left and right pial surfaces (as GIFTI files)
ref_file – Reference image (one 3D volume) to define the target space
- Outputs:
anat_ribbon – Cortical gray matter mask, sampled into
ref_filespace
- smriprep.workflows.surfaces.init_autorecon_resume_wf(*, omp_nthreads, name='autorecon_resume_wf')[source]
Resume recon-all execution, assuming the -autorecon1 stage has been completed.
In order to utilize resources efficiently, this is broken down into seven sub-stages; after the first stage, the second and third stages may be run simultaneously, and the fifth and sixth stages may be run simultaneously, if resources permit; the fourth stage must be run prior to the fifth and sixth, and the seventh must be run after:
$ recon-all -sd <output dir>/freesurfer -subjid sub-<subject_label> \ -autorecon2-volonly $ recon-all -sd <output dir>/freesurfer -subjid sub-<subject_label> \ -autorecon-hemi lh -T2pial \ -noparcstats -noparcstats2 -noparcstats3 -nohyporelabel -nobalabels $ recon-all -sd <output dir>/freesurfer -subjid sub-<subject_label> \ -autorecon-hemi rh -T2pial \ -noparcstats -noparcstats2 -noparcstats3 -nohyporelabel -nobalabels $ recon-all -sd <output dir>/freesurfer -subjid sub-<subject_label> \ -cortribbon $ recon-all -sd <output dir>/freesurfer -subjid sub-<subject_label> \ -autorecon-hemi lh -nohyporelabel $ recon-all -sd <output dir>/freesurfer -subjid sub-<subject_label> \ -autorecon-hemi rh -nohyporelabel $ recon-all -sd <output dir>/freesurfer -subjid sub-<subject_label> \ -autorecon3The parcellation statistics steps are excluded from the second and third stages, because they require calculation of the cortical ribbon volume (the fourth stage). Hypointensity relabeling is excluded from hemisphere-specific steps to avoid race conditions, as it is a volumetric operation.
- Workflow Graph
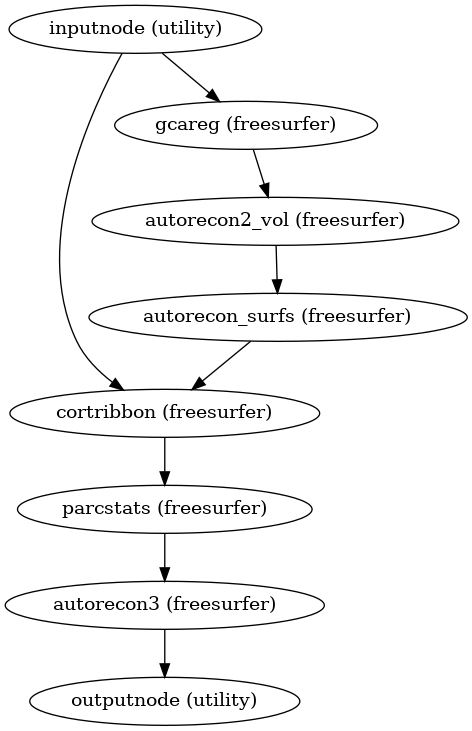
(Source code, png, svg, pdf)
- Inputs:
subjects_dir – FreeSurfer SUBJECTS_DIR
subject_id – FreeSurfer subject ID
use_T2 – Refine pial surface using T2w image
use_FLAIR – Refine pial surface using FLAIR image
- Outputs:
subjects_dir – FreeSurfer SUBJECTS_DIR
subject_id – FreeSurfer subject ID
- smriprep.workflows.surfaces.init_cortex_masks_wf(*, name: str = 'cortex_masks_wf')[source]
Create cortical surface masks from surface files.
- Workflow Graph
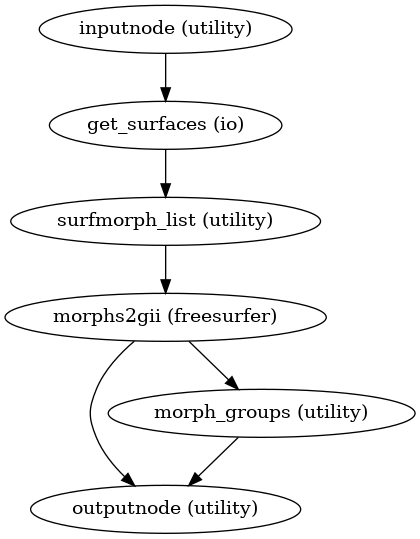
(Source code, png, svg, pdf)
- Inputs:
midthickness (len-2 list of str) – Each hemisphere’s FreeSurfer midthickness surface file in GIFTI format
thickness (len-2 list of str) – Each hemisphere’s FreeSurfer thickness file in GIFTI format
- Outputs:
cortex_masks (len-2 list of str) – Cortical surface mask in GIFTI format for each hemisphere
source_files (len-2 list of lists of str) – Each hemisphere’s source files, which are used to create the mask
- smriprep.workflows.surfaces.init_fsLR_reg_wf(*, name='fsLR_reg_wf')[source]
Generate GIFTI registration files to fsLR space
- smriprep.workflows.surfaces.init_gifti_morphometrics_wf(*, morphometrics: list[str] = ('thickness', 'curv', 'sulc'), name: str = 'gifti_morphometrics_wf')[source]
Prepare GIFTI shape files from morphometrics found in a FreeSurfer subjects directory.
The default morphometrics are
lh/rh.thickness,lh/rh.curv, andlh/rh.sulc.- Workflow Graph
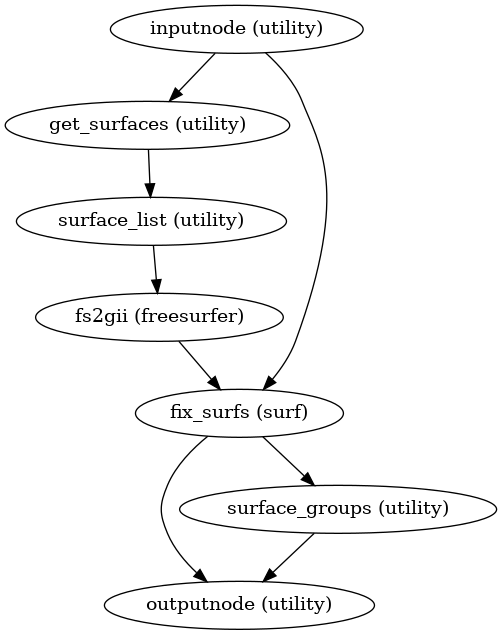
(Source code, png, svg, pdf)
- Inputs:
subjects_dir – FreeSurfer SUBJECTS_DIR
subject_id – FreeSurfer subject ID
- Outputs:
morphometrics – GIFTI shape files for all requested morphometrics
``<morphometric>`` – Left and right GIFTIs for each morphometry type passed to
morphometrics
- smriprep.workflows.surfaces.init_gifti_surfaces_wf(*, surfaces: list[str] = ('pial', 'midthickness', 'inflated', 'white'), to_scanner: bool = True, name: str = 'gifti_surface_wf')[source]
Prepare GIFTI surfaces from a FreeSurfer subjects directory.
The default surfaces are
lh/rh.pial,lh/rh.midthickness,lh/rh.inflated, andlh/rh.white.Vertex coordinates are
transformedto align with native anatomical space whenfsnative2anat_xfmis provided.- Workflow Graph
(Source code, png, svg, pdf)
- Inputs:
subjects_dir – FreeSurfer SUBJECTS_DIR
subject_id – FreeSurfer subject ID
fsnative2anat_xfm – LTA formatted affine transform file
- Outputs:
surfaces – GIFTI surfaces for all requested surfaces
``<surface>`` – Left and right GIFTIs for each surface passed to
surfaces
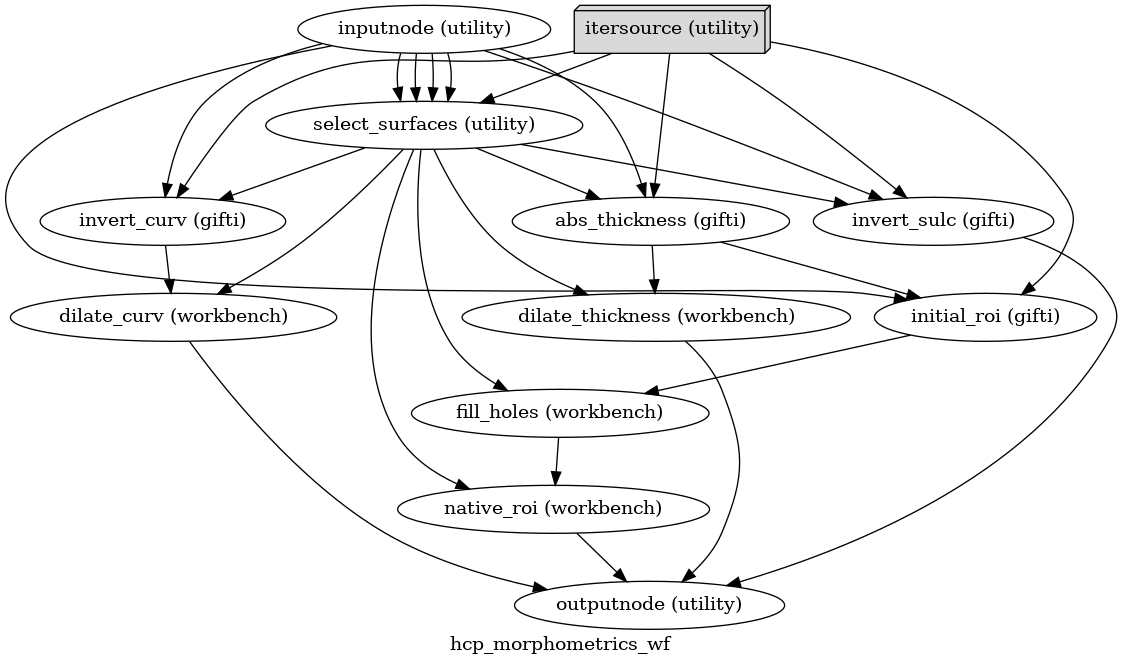
- smriprep.workflows.surfaces.init_hcp_morphometrics_wf(*, omp_nthreads: int, name: str = 'hcp_morphometrics_wf') LiterateWorkflow[source]
Convert FreeSurfer-style morphometrics to HCP-style.
The HCP uses different conventions for curvature and sulcal depth from FreeSurfer, and performs some geometric preprocessing steps to get final metrics and a subject-specific cortical ROI.
Workflow Graph
(Source code, png, svg, pdf)
- Parameters:
omp_nthreads – Maximum number of threads an individual process may use
- Inputs:
subject_id – FreeSurfer subject ID
thickness – FreeSurfer thickness file in GIFTI format
curv – FreeSurfer curvature file in GIFTI format
sulc – FreeSurfer sulcal depth file in GIFTI format
- Outputs:
thickness – HCP-style thickness file in GIFTI format
curv – HCP-style curvature file in GIFTI format
sulc – HCP-style sulcal depth file in GIFTI format
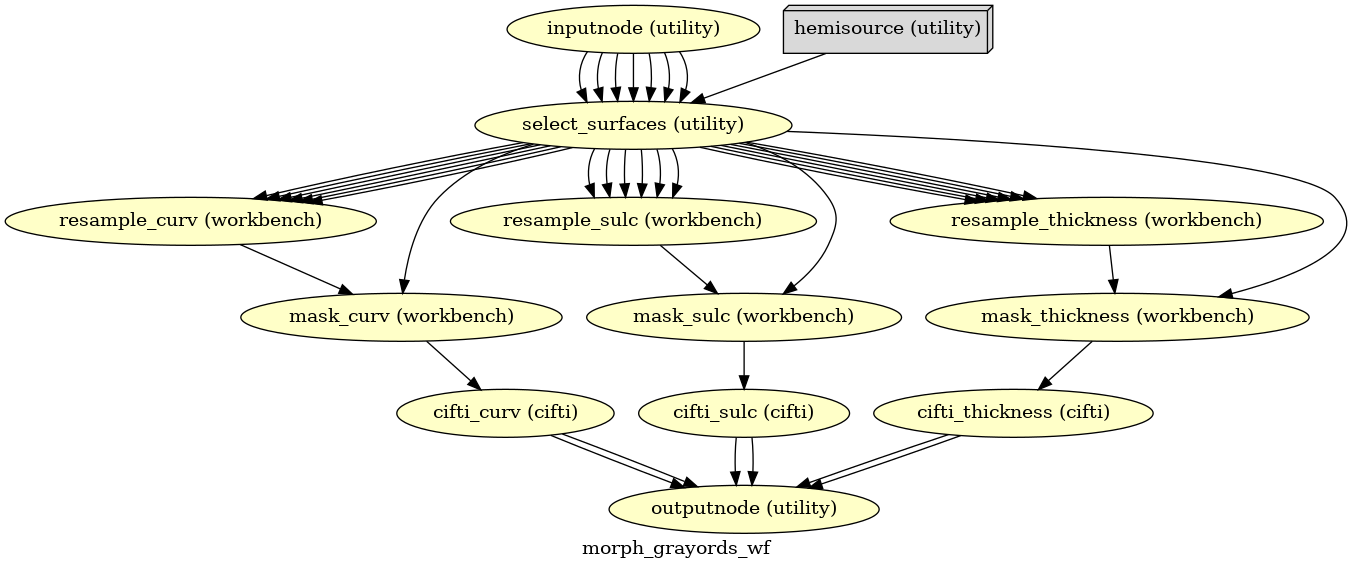
- smriprep.workflows.surfaces.init_morph_grayords_wf(grayord_density: Literal['91k', '170k'], omp_nthreads: int, name: str = 'morph_grayords_wf')[source]
Sample Grayordinates files onto the fsLR atlas.
Outputs are in CIFTI2 format.
- Workflow Graph
(Source code, png, svg, pdf)
- Parameters:
- Inputs:
curv – HCP-style curvature file in GIFTI format
sulc – HCP-style sulcal depth file in GIFTI format
thickness – HCP-style thickness file in GIFTI format
roi – HCP-style cortical ROI file in GIFTI format
midthickness – GIFTI surface mesh corresponding to the midthickness surface
sphere_reg_fsLR – GIFTI surface mesh corresponding to the subject’s fsLR registration sphere
- Outputs:
curv_fsLR – HCP-style curvature file in CIFTI-2 format, resampled to fsLR
curv_metadata – Path to JSON file containing metadata corresponding to
curv_fsLRsulc_fsLR – HCP-style sulcal depth file in CIFTI-2 format, resampled to fsLR
sulc_metadata – Path to JSON file containing metadata corresponding to
sulc_fsLRthickness_fsLR – HCP-style thickness file in CIFTI-2 format, resampled to fsLR
- smriprep.workflows.surfaces.init_msm_sulc_wf(*, sloppy: bool = False, name: str = 'msm_sulc_wf')[source]
Run MSMSulc registration to fsLR surfaces, per hemisphere.
- smriprep.workflows.surfaces.init_refinement_wf(*, image_type: Literal['T1w', 'T2w'] = 'T1w', name: str = 'refinement_wf') LiterateWorkflow[source]
Refine ANTs brain extraction with FreeSurfer segmentation
- Workflow Graph
(Source code, png, svg, pdf)
- Inputs:
subjects_dir – FreeSurfer SUBJECTS_DIR
subject_id – FreeSurfer subject ID
fsnative2anat_xfm – LTA-style affine matrix translating from FreeSurfer-conformed subject space to anatomical
reference_image – Input
ants_segs – Brain tissue segmentation from ANTS
antsBrainExtraction.shsubjects_dir – FreeSurfer SUBJECTS_DIR
subject_id – FreeSurfer subject ID
- Outputs:
out_brainmask – Refined brainmask, derived from FreeSurfer’s
asegvolume
See also
- smriprep.workflows.surfaces.init_resample_surfaces_wf(surfaces: list[str], template: str = 'fsLR', cohort: str | None = None, space: str | None = 'fsLR', density: str | None = None, grayord_density: str | None = None, name: str = 'resample_surfaces_wf')[source]
Resample subject surfaces surface to specified template space and density, using a given registration space.
- Workflow Graph
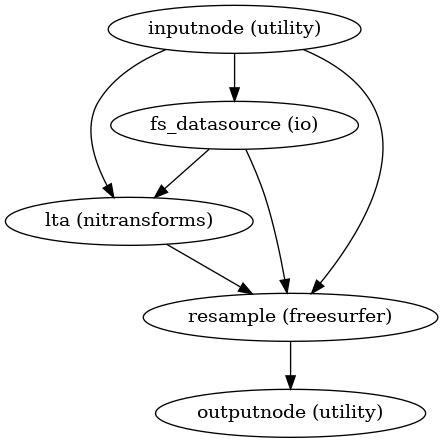
(Source code, png, svg, pdf)
- Parameters:
surfaces (
listofstr) – Names of surfaces (e.g.,'white') to resample. Both hemispheres will be resampled.template (
str) – The template space to resample to, e.g.,'onavg','fsLR'.cohort (
strorNone) – The template cohort to use, if the template provides multiple.space (
strorNone) – The registration space for which there are both subject and template registration spheres. IfNone, the template space is used.density (
str) – The density to resample to, e.g.,'10k','41k'. Number of vertices per hemisphere.name (
str) – Unique name for the subworkflow (default:"resample_surfaces_wf")
- Inputs:
``<surface>`` – Left and right GIFTIs for each surface name passed to
surfaces.``sphere_reg_<space>`` – GIFTI surface mesh corresponding to the subject’s registration sphere to
space.
- Outputs:
``<surface>_<template>`` – Left and right GIFTI surface mesh corresponding to the input surface, resampled to the specified space and density.
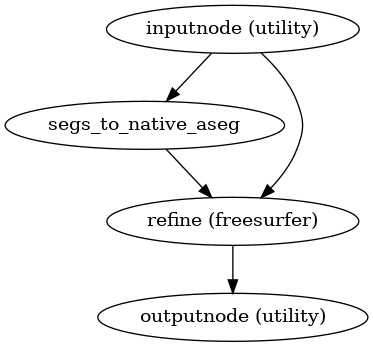
- smriprep.workflows.surfaces.init_segs_to_native_wf(*, image_type: Literal['T1w', 'T2w'] = 'T1w', segmentation: Literal['aseg', 'aparc_aseg', 'aparc_a2009s', 'aparc_dkt'] | str = 'aseg', name: str = 'segs_to_native_wf') LiterateWorkflow[source]
Get a segmentation from FreeSurfer conformed space into native anatomical space.
- Workflow Graph
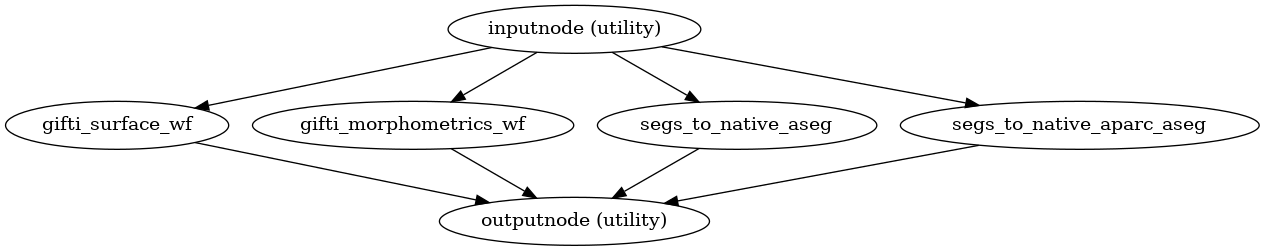
(Source code, png, svg, pdf)
- Parameters:
image_type – MR anatomical image type (‘T1w’ or ‘T2w’)
segmentation – The name of a segmentation (‘aseg’ or ‘aparc_aseg’ or ‘wmparc’)
- Inputs:
in_file – Anatomical, merged anatomical image after INU correction
subjects_dir – FreeSurfer SUBJECTS_DIR
subject_id – FreeSurfer subject ID
fsnative2anat_xfm – LTA-style affine matrix translating from FreeSurfer-conformed subject space to anatomical
- Outputs:
out_file – The selected segmentation, after resampling in native space
- smriprep.workflows.surfaces.init_surface_derivatives_wf(*, image_type: Literal['T1w', 'T2w'] = 'T1w', name: str = 'surface_derivatives_wf')[source]
Generate sMRIPrep derivatives from FreeSurfer derivatives
- Workflow Graph
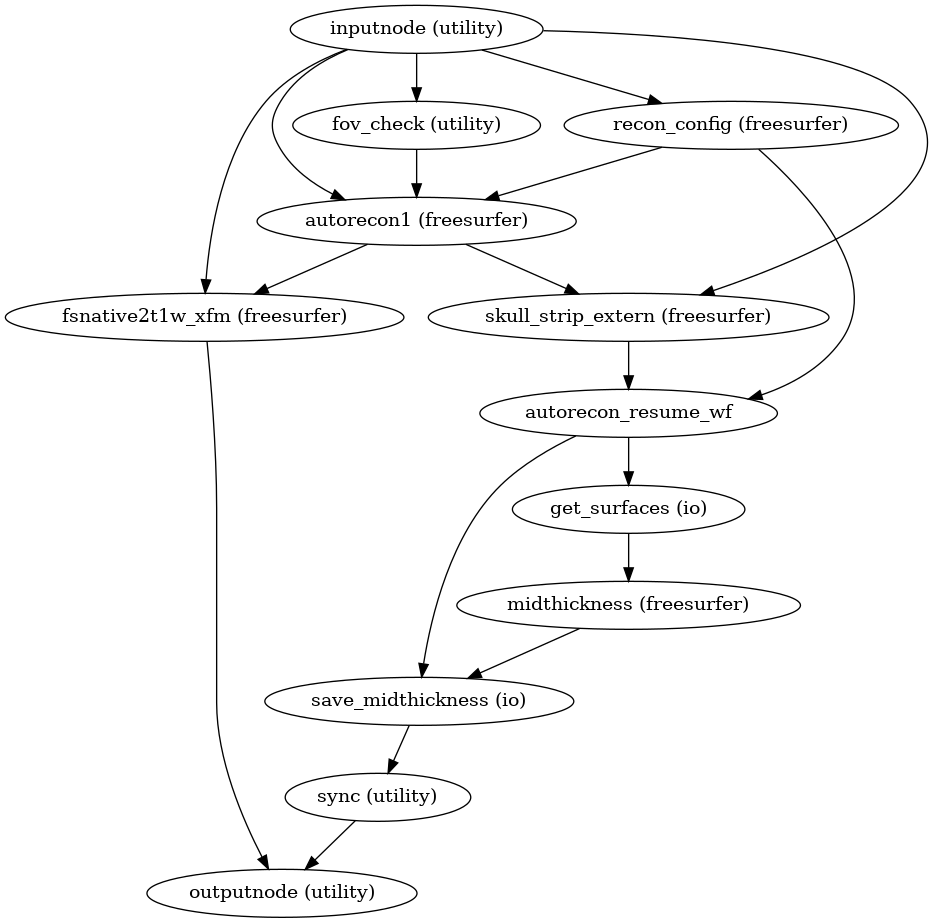
(Source code, png, svg, pdf)
- Inputs:
reference – Reference image in native anatomical space, for defining a resampling grid
fsnative2anat_xfm – LTA-style affine matrix translating from FreeSurfer-conformed subject space to anatomical
subjects_dir – FreeSurfer SUBJECTS_DIR
subject_id – FreeSurfer subject ID
- Outputs:
surfaces – GIFTI surfaces for gray/white matter boundary, pial surface, midthickness (or graymid) surface, and inflated surfaces
morphometrics – GIFTIs of cortical thickness, curvature, and sulcal depth
out_aseg – FreeSurfer’s aseg segmentation, in native anatomical space
out_aparc – FreeSurfer’s aparc+aseg segmentation, in native anatomical space
See also
init_gifti_surface_wf()
- smriprep.workflows.surfaces.init_surface_recon_wf(*, omp_nthreads: int, hires: bool, fs_no_resume: bool, precomputed: dict, name='surface_recon_wf')[source]
Reconstruct anatomical surfaces using FreeSurfer’s
recon-all.Reconstruction is performed in three phases. The first phase initializes the subject with T1w and T2w (if available) structural images and performs basic reconstruction (
autorecon1) with the exception of skull-stripping. For example, a subject with only one session with T1w and T2w images would be processed by the following command:$ recon-all -sd <output dir>/freesurfer -subjid sub-<subject_label> \ -i <bids-root>/sub-<subject_label>/anat/sub-<subject_label>_T1w.nii.gz \ -T2 <bids-root>/sub-<subject_label>/anat/sub-<subject_label>_T2w.nii.gz \ -autorecon1 \ -noskullstrip -noT2pial -noFLAIRpialThe second phase imports an externally computed skull-stripping mask. This workflow refines the external brainmask using the internal mask implicit the the FreeSurfer’s
aseg.mgzsegmentation, to reconcile ANTs’ and FreeSurfer’s brain masks.First, the
aseg.mgzmask from FreeSurfer is refined in two steps, using binary morphological operations:With a binary closing operation the sulci are included into the mask. This results in a smoother brain mask that does not exclude deep, wide sulci.
Fill any holes (typically, there could be a hole next to the pineal gland and the corpora quadrigemina if the great cerebral brain is segmented out).
Second, the brain mask is grown, including pixels that have a high likelihood to the GM tissue distribution:
Dilate and subtract the brain mask, defining the region to search for candidate pixels that likely belong to cortical GM.
Pixels found in the search region that are labeled as GM by ANTs (during
antsBrainExtraction.sh) are directly added to the new mask.Otherwise, estimate GM tissue parameters locally in patches of
wwsize, and test the likelihood of the pixel to belong in the GM distribution.
This procedure is inspired on mindboggle’s solution to the problem: https://github.com/nipy/mindboggle/blob/7f91faaa7664d820fe12ccc52ebaf21d679795e2/mindboggle/guts/segment.py#L1660
The final phase resumes reconstruction, using the T2w image to assist in finding the pial surface, if available. See
init_autorecon_resume_wf()for details.Memory annotations for FreeSurfer are based off their documentation. They specify an allocation of 4GB per subject. Here we define 5GB to have a certain margin.
- Workflow Graph

(Source code, png, svg, pdf)
- Parameters:
omp_nthreads (int) – Maximum number of threads an individual process may use
hires (bool) – Enable sub-millimeter preprocessing in FreeSurfer
fs_no_resume (bool) – use precomputed freesurfer without attempting to resume (eg. for longitudinal base or fastsurfer)
- Inputs:
t1w – List of T1-weighted structural images
t2w – List of T2-weighted structural images (only first used)
flair – List of FLAIR images
skullstripped_t1 – Skull-stripped T1-weighted image (or mask of image)
subjects_dir – FreeSurfer SUBJECTS_DIR
subject_id – FreeSurfer subject ID
- Outputs:
subjects_dir – FreeSurfer SUBJECTS_DIR
subject_id – FreeSurfer subject ID
fsnative2t1w_xfm – LTA-style affine matrix translating from FreeSurfer-conformed subject space to T1w
See also