smriprep.workflows.anatomical module
Anatomical reference preprocessing workflows.
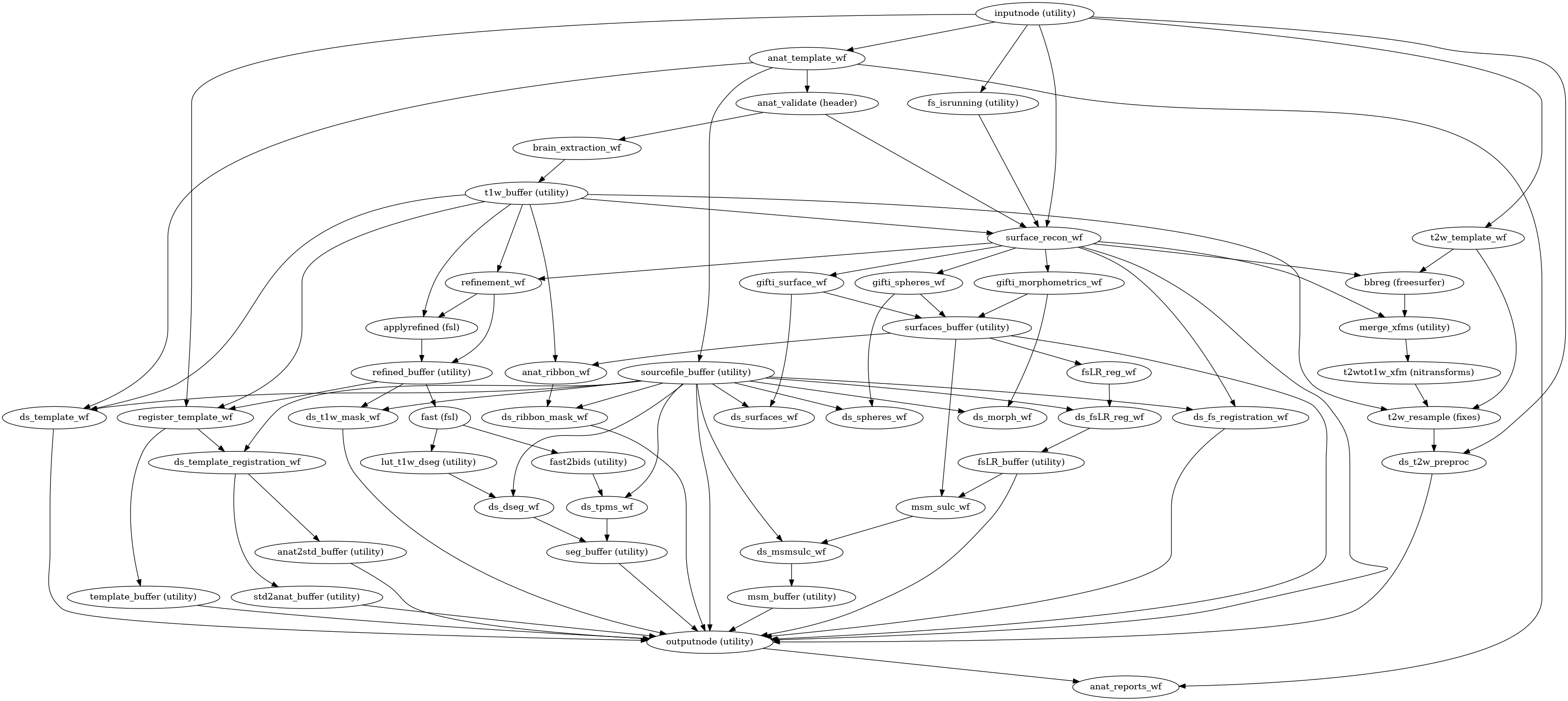
- smriprep.workflows.anatomical.init_anat_fit_wf(*, bids_root: str, output_dir: str, freesurfer: bool, hires: bool, longitudinal: bool, msm_sulc: bool, t1w: list, t2w: list, skull_strip_mode: str, skull_strip_template: Reference, spaces: SpatialReferences, precomputed: dict, omp_nthreads: int, flair: list = (), debug: bool = False, sloppy: bool = False, name='anat_fit_wf', skull_strip_fixed_seed: bool = False, fs_no_resume: bool = False)[source]
Stage the anatomical preprocessing steps of sMRIPrep.
This includes:
T1w reference: realigning and then averaging T1w images.
Brain extraction and INU (bias field) correction.
Brain tissue segmentation.
Spatial normalization to standard spaces.
Surface reconstruction with FreeSurfer.
- Workflow Graph
(Source code, png, svg, pdf)
- Parameters:
bids_root (
str) – Path of the input BIDS dataset rootoutput_dir (
str) – Directory in which to save derivativesfreesurfer (
bool) – Enable FreeSurfer surface reconstruction (increases runtime by 6h, at the very least)hires (
bool) – Enable sub-millimeter preprocessing in FreeSurferlongitudinal (
bool) – Create unbiased structural template, regardless of number of inputs (may increase runtime)t1w (
list) – List of T1-weighted structural images.skull_strip_mode (
str) – Determiner for T1-weighted skull stripping (force ensures skull stripping, skip ignores skull stripping, and auto automatically ignores skull stripping if pre-stripped brains are detected).skull_strip_template (
Reference) – Spatial reference to use in atlas-based brain extraction.spaces (
SpatialReferences) – Object containing standard and nonstandard space specifications.precomputed (
dict) – Dictionary mapping output specification attribute names and paths to precomputed derivatives.omp_nthreads (
int) – Maximum number of threads an individual process may usedebug (
bool) – Enable debugging outputssloppy (
bool) – Quick, impercise operations. Used to decrease workflow duration.name (
str, optional) – Workflow name (default: anat_fit_wf)skull_strip_fixed_seed (
bool) – Do not use a random seed for skull-stripping - will ensure run-to-run replicability when used with –omp-nthreads 1 (default:False).
- Inputs:
t1w – List of T1-weighted structural images
t2w – List of T2-weighted structural images
roi – A mask to exclude regions during standardization
flair – List of FLAIR images
subjects_dir – FreeSurfer SUBJECTS_DIR
subject_id – FreeSurfer subject ID
- Outputs:
t1w_preproc – The T1w reference map, which is calculated as the average of bias-corrected and preprocessed T1w images, defining the anatomical space.
t1w_mask – Brain (binary) mask estimated by brain extraction.
t1w_dseg – Brain tissue segmentation of the preprocessed structural image, including gray-matter (GM), white-matter (WM) and cerebrospinal fluid (CSF).
t1w_tpms – List of tissue probability maps corresponding to
t1w_dseg.t1w_valid_list – List of input T1w images accepted for preprocessing. If t1w_preproc is precomputed, this is always a list containing that image.
template – List of template names to which the structural image has been registered
anat2std_xfm – List of nonlinear spatial transforms to resample data from subject anatomical space into standard template spaces. Collated with template.
std2anat_xfm – List of nonlinear spatial transforms to resample data from standard template spaces into subject anatomical space. Collated with template.
subjects_dir – FreeSurfer SUBJECTS_DIR; use as input to a node to ensure that it is run after FreeSurfer reconstruction is completed.
subject_id – FreeSurfer subject ID; use as input to a node to ensure that it is run after FreeSurfer reconstruction is completed.
fsnative2t1w_xfm – ITK-style affine matrix translating from FreeSurfer-conformed subject space to T1w
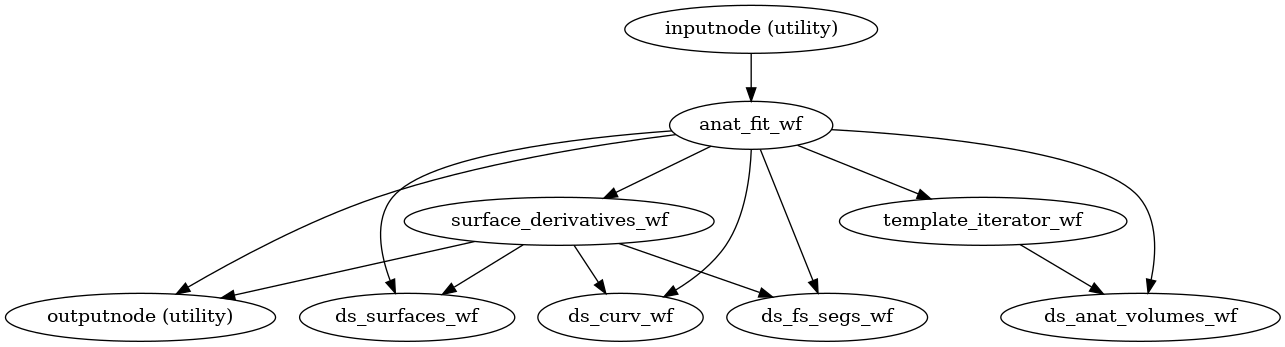
- smriprep.workflows.anatomical.init_anat_preproc_wf(*, bids_root: str, output_dir: str, freesurfer: bool, hires: bool, longitudinal: bool, msm_sulc: bool, t1w: list, t2w: list, skull_strip_mode: str, skull_strip_template: Reference, spaces: SpatialReferences, precomputed: dict, omp_nthreads: int, flair: list = (), debug: bool = False, sloppy: bool = False, cifti_output: Literal['91k', '170k', False] = False, name: str = 'anat_preproc_wf', skull_strip_fixed_seed: bool = False, fs_no_resume: bool = False)[source]
Stage the anatomical preprocessing steps of sMRIPrep.
This workflow is a compatibility wrapper around
init_anat_fit_wf()that emits all derivatives that were present in sMRIPrep 0.9.x and before.- Workflow Graph
(Source code, png, svg, pdf)
- Parameters:
bids_root (
str) – Path of the input BIDS dataset rootoutput_dir (
str) – Directory in which to save derivativesfreesurfer (
bool) – Enable FreeSurfer surface reconstruction (increases runtime by 6h, at the very least)hires (
bool) – Enable sub-millimeter preprocessing in FreeSurferlongitudinal (
bool) – Create unbiased structural template, regardless of number of inputs (may increase runtime)t1w (
list) – List of T1-weighted structural images.skull_strip_mode (
str) – Determiner for T1-weighted skull stripping (force ensures skull stripping, skip ignores skull stripping, and auto automatically ignores skull stripping if pre-stripped brains are detected).skull_strip_template (
Reference) – Spatial reference to use in atlas-based brain extraction.spaces (
SpatialReferences) – Object containing standard and nonstandard space specifications.precomputed (
dict) – Dictionary mapping output specification attribute names and paths to precomputed derivatives.omp_nthreads (
int) – Maximum number of threads an individual process may usedebug (
bool) – Enable debugging outputssloppy (
bool) – Quick, impercise operations. Used to decrease workflow duration.name (
str, optional) – Workflow name (default: anat_fit_wf)skull_strip_fixed_seed (
bool) – Do not use a random seed for skull-stripping - will ensure run-to-run replicability when used with –omp-nthreads 1 (default:False).fs_no_resume (bool) – EXPERT: Import pre-computed FreeSurfer reconstruction without resuming. The user is responsible for ensuring that all necessary files are present. (default:
False).
- Inputs:
t1w – List of T1-weighted structural images
t2w – List of T2-weighted structural images
roi – A mask to exclude regions during standardization
flair – List of FLAIR images
subjects_dir – FreeSurfer SUBJECTS_DIR
subject_id – FreeSurfer subject ID
- Outputs:
t1w_preproc – The T1w reference map, which is calculated as the average of bias-corrected and preprocessed T1w images, defining the anatomical space.
t1w_mask – Brain (binary) mask estimated by brain extraction.
t1w_dseg – Brain tissue segmentation of the preprocessed structural image, including gray-matter (GM), white-matter (WM) and cerebrospinal fluid (CSF).
t1w_tpms – List of tissue probability maps corresponding to
t1w_dseg.template – List of template names to which the structural image has been registered
anat2std_xfm – List of nonlinear spatial transforms to resample data from subject anatomical space into standard template spaces. Collated with template.
std2anat_xfm – List of nonlinear spatial transforms to resample data from standard template spaces into subject anatomical space. Collated with template.
subjects_dir – FreeSurfer SUBJECTS_DIR; use as input to a node to ensure that it is run after FreeSurfer reconstruction is completed.
subject_id – FreeSurfer subject ID; use as input to a node to ensure that it is run after FreeSurfer reconstruction is completed.
fsnative2t1w_xfm – ITK-style affine matrix translating from FreeSurfer-conformed subject space to T1w
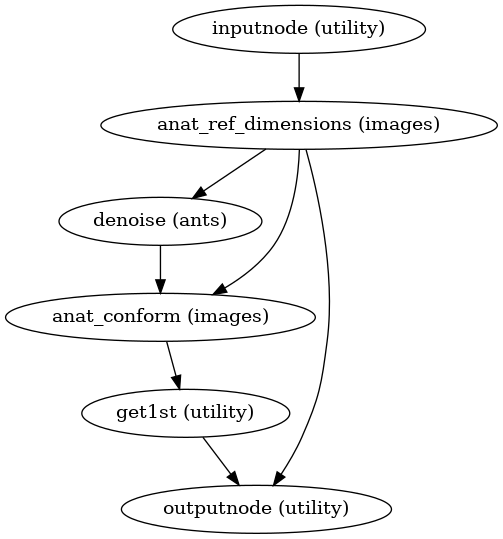
- smriprep.workflows.anatomical.init_anat_template_wf(*, longitudinal: bool, omp_nthreads: int, num_files: int, image_type: Literal['T1w', 'T2w'], name: str = 'anat_template_wf')[source]
Generate a canonically-oriented, structural average from all input images.
- Workflow Graph
(Source code, png, svg, pdf)
- Parameters:
longitudinal (
bool) – Create unbiased structural average, regardless of number of inputs (may increase runtime)omp_nthreads (
int) – Maximum number of threads an individual process may usenum_files (
int) – Number of imagesimage_type (
str) – MR image type (T1w, T2w, etc.)name (
str, optional) – Workflow name (default: anat_template_wf)
- Inputs:
anat_files – List of structural images
- Outputs:
anat_ref – Structural reference averaging input images
anat_valid_list – List of structural images accepted for combination
anat_realign_xfm – List of affine transforms to realign input images to final reference
out_report – Conformation report